AlphaFold2 and Coronavirus
“We cannot solve our problems with the same thinking we used when we created them.” - Albert Einstein
Introduction
In late December 2021, during an internship at NyBerMan Merit from LLBio-IT School, I embarked on a project to predict Coronavirus glycoprotein structures using AlphaFold2. This project was not originally part of the internship’s pipeline, but due to my interest and initiative, I decided to take it on independently. The further implementation of this project was done in my free time after the internship at the end of April 2022 - beginning of May 2022.
Progress
Initial Steps
After the internship skipped AlphaFold due to time constraints, I researched and found tutorials that helped me set up my own analysis. I initially struggled with system requirements but overcame them by leveraging cloud solutions. The most helpful resource was the blog and notebooks of Colby T. Ford, which provided a detailed guide on using AlphaFold2 for protein structure prediction on Azure Machine Learning.
Data Collection
I focused on collecting polymorphism data from various strains of the virus. This included identifying crucial sequences in the Spike protein’s receptor binding domain, which was critical for accurate modeling. To my joy, the data was already prepared throughout the internship, so I could start the analysis right away.
Setting Up Azure ML and Jupyter

I used Jupyter notebooks for scripting and Azure Machine Learning for building the program. The initial setup involved installing necessary configurations and writing a run script based on existing templates. Some challenges arose due to the complexity of the AlphaFold2 model, but I managed to resolve them by following the tutorials. Also, I’ve never used Azure Machine Learning or any other cloud-based ML platform before, so it was a great learning experience (after everythong finally worked out :).
Experiment Configuration and Execution
I configured the Azure cluster, set up experiment scripts, and defined dependencies using a Docker image. The HyperDrive feature was used to fine-tune the model parameters, after which I ran the experiment and logged all necessary data. The process was time-consuming but rewarding, as I could see the model’s progress and make adjustments as needed.
Results
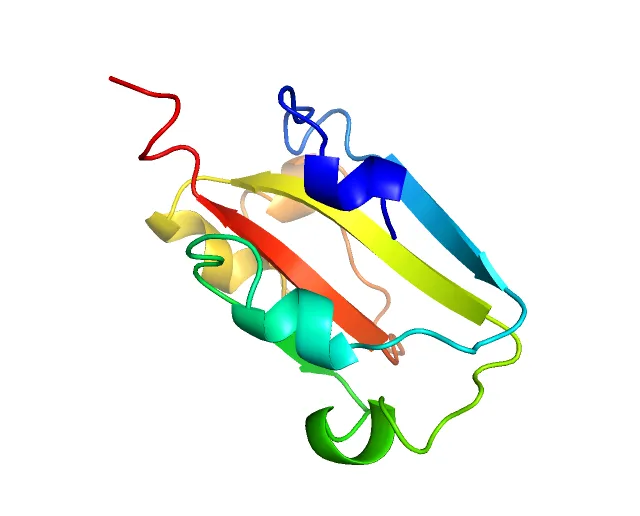
The experiment successfully predicted the structure of the Spike protein’s receptor binding domain. The results were visualized using 3Dmol, providing a clear view of the protein’s configuration. The model’s accuracy was impressive, with the predicted structure closely matching the actual protein. The detailed analysis and visualization steps are available in my GitHub repository. The image below shows the structure of the Omicron variant’s Spike protein, which was accurately predicted by the model for one part of the strains.
Technology Learned/Used
Throughout this project, I deepened my understanding of:
- AlphaFold2 for protein structure prediction
- Azure Machine Learning for managing and executing ML experiments
- Docker for environment management
- Jupyter and Python for scripting and data handling
Conclusion
The fascinating ability of AlphaFold2 to predict protein structures has opened up new possibilities for understanding complex biological systems. Through this project, I gained valuable experience in using this cutting-edge technology to solve real-world problem: predicting the structure of proteins (Coronavirus glycoprotein in my case). The results were promising, and I look forward to further exploring the potential applications of AlphaFold2 in the field of bioinformatics.
